By Raffaele Conte
Introduction
On 29 December 2019, Chinese authorities identified a cluster of similar pneumonia cases of unknown aetiology in Wuhan City, Hubei Province, China and a novel strain of coronavirus (2019-nCoV) was subsequently isolated from a patient on 7 January 2020.1 The source of the virus is unknown, also unknown is if most cases from the initial cluster had epidemiological links with a live animal market (Huanan South China Seafood Market), suggesting a possible zoonotic origin. Despite this, the occurrence of human-to-human transmission was confirmed by infections in family, social clusters, and in healthcare workers through respiratory droplets.2,3 The World Health Organisation (WHO) on 11 March 2020 declared the novel coronavirus disease (COVID-19) a global pandemic, and currently the 2019-nCoV is diffused all over the world, especially USA and Europe (see table 1).
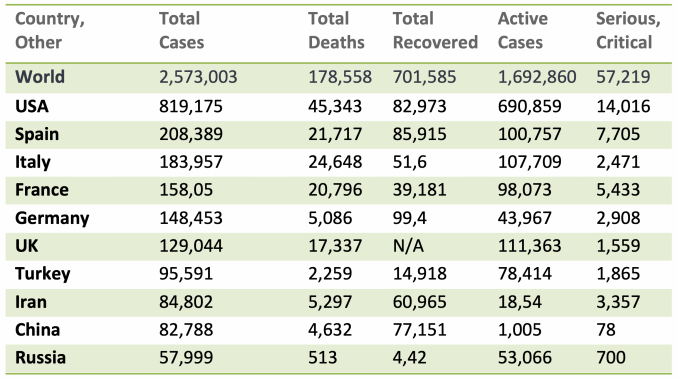 Table 1. COVID-19 spread. Data of 22 April 2020. (Credit https://www.worldometers.info/coronavirus/)
Table 1. COVID-19 spread. Data of 22 April 2020. (Credit https://www.worldometers.info/coronavirus/)
The severity of these data, combined with the experience of the three outbreaks of human coronavirus (HCoV) infections at the beginning of the twenty century (Spanish flu) and of the twenty-first century (SARS-Severe acute respiratory syndrome 2002, and MERS-Middle East respiratory syndrome 2012) have highlighted the necessity for readily available, accurate and fast diagnostic testing methods.
Laboratory technicians are considered by the governments of both UK and Italy as workers “essential” to counteracting the COVID-19 pandemic due to their primary role in patient assistance and 2019-nCoV diagnosis. This article analyses the role of technicians in specimen collection and genetic analysis of 2019-nCoV strain and highlights the importance of an effective collaboration between the public and private health systems through the case study of the AMES diagnostic centre in its activity for the GENCOVID project in association with the experimental Zooprophylactic institute for the Campania region (figure 1).
 Figure 1. GENCOVID project at AMES polydiagnostic center
Figure 1. GENCOVID project at AMES polydiagnostic center
Role of technicians in specimen collections
The reliability of laboratory testing methods for 2019-nCoV (also called SARS-CoV-2) depends on the collection of the correct specimen from the patient at the right time. In particular, the endemic SARS-CoV-2 was detected in upper and lower respiratory sources including throat, nasal nasopharyngeal (NP), sputum, and bronchial fluid.4, 5 Wan et al reported that oropharyngeal (OP) swabs were used much more frequently than NP swabs in China during the COVID19 outbreak; however, the 2019-nCoV RNA was detected only in 32% of OP swabs, a value which was significantly lower than that in NP swabs (63%).6 Because of this, current indications for SARS-CoV-2 testing recommend to collect the upper respiratory NP swab or both upper and lower respiratory samples in the same tube.7However, this type of collection (especially if it is produced via sputum or via bronchoscopy) increases biosafety risk to healthcare workers because of the creation of aerosol droplets. It therefore requires a proper use of personal protective equipment (PPE). Specimens collected for laboratory testing of 2019-nCoV should be maintained at refrigerated temperature for up to 72 h, or frozen at −70°C or below.
Role of technicians in the molecular analysis
Random-amplification deep-sequencing approaches are the gold standard for identifying SARS-CoV-2 in OP\NP swabs.8, 9 For clinical diagnostic applications, the genetic heterogeneity of HCoVs precludes a single “pan-HCoV” molecular assay.10 In fact, some pan-CoV assays use degenerate primers,11 some employ a single set of nondegenerate primers,12 while others utilize multiple primer sets13 like the current molecular respiratory panel detecting the endemic HCoVs (HCoV-NL63, HCoV-HKU1, HCoV-OC43, and HCoV-229E) that uses multiple sets of PCR oligonucleotides.14, 15 SARS-CoV-2 tests are made according to the US CDC RT-PCR Diagnostic Panel for universal detection of SARS-like beta-coronaviruses.16 In particular, three separate RT-PCR reactions target the N gene. One primer set detects all beta-coronaviruses, while two sets are specific for SARS-CoV-2. All 3 assays must be positive to report presumptive positive for 2019-nCoV. The instruments used in AMES for SARS-CoV-2 RNA retro-transcription and cDNA amplification are: four robot Hamilton for the automation of the extraction, 2 robot NIPT, 2 robot hexome, 7 next generation sequencers (NGS; in particular 2 Miseq, 4 Nextseq, 1 Novaseq 6000) and 1 iSCAN reader. All these instruments permit to potentially analyse 2000 samples a day, and are certified by Illumina. Laboratory technicians have a fundamental role in the actuation of analysis. In particular the first passage, that is the inactivation of the virus requiring technicians to aliquot the samples and add in a volumetric ratio 1:1 the inactivation buffer. This is a saline solution prepared to induce 2019-nCoV lysis, in order to extract RNA. This is the most dangerous part of the process due to the fact that, for positive samples, the SARS-CoV-2 can still be alive. Furthermore, contaminations must be avoided. Consequently, such a process must be done in a biological hood of level 2 of safety (a laminar flow hood). In addition, the area of the laboratory in which inactivation, extraction and amplification of 2019-nCoV genetic material is done must be organised such to be separated from the other zones, and staff must wear full body single use garments for clean room environments. Once the procedure is completed, such areas must be sterilised with ultraviolet waves (UV) and ozone. Fig. 2 shows the area of AMES polydiagnostic center designated for SARS-CoV-2 analysis.
 Fig. 2. Area designated for the analysis of SARS-CoV-2 in AMES polydiagnostic center
Fig. 2. Area designated for the analysis of SARS-CoV-2 in AMES polydiagnostic center
Samples are then placed on the extractor (or alternatively, hand extracted by adding isopropanol and a high salt solution before centrifugation, in order to pellet the RNA). The extracted material is retro-amplificated by PCR using the automatic sequencers available in AMES. If viral RNA is retro-transcribed into cDNA the sample is positive, on the other hand it is negative. Post analytical validation of results must be done by highly specialised technicians with experience in genetic and molecular biology.
Data from AMES polydiagnostic centre
The activity of AMES polydiagnostic centre against the COVID-19 pandemic acts as example of the efficient public-private collaboration in the Italian health sector. The crisis unit of the Campania region organised a network of 12 public laboratories for the analysis of samples. These have been assisted by private laboratories, including the AMES polydiagnostic centre, to manage the enormous amount of work. The AMES series began on 4 April 2020 and reached, on 25 April 2020, about 9000 samples. The percentage of positives fell from 3% of the first week to 0.59% at 25 April 2020, highlighting the effective containment action of the regional lockdown. The statistic accuracy of these data is strengthened by the large number of tests carried out: 9000 out of 65000, equal to 13.85% of the Campania region. Another particular to highlight is the model of free collaboration given by the AMES polydiagnostic centre to the Italian health services. In fact, genetic testing material was provided by Campania region while the private centre provided equipment and competent staff as well as financial aid, useful to overcome this emergency situation.
Conclusion
This article highlights the fundamental role of technicians in emergency situations. Having trained technical and specialist staff constitutes a solid basis to place procedures aimed to overcome complicated situations. Similarly, a strong cooperation between private and public structures help to share competencies and productivity for faster decisions.
Author

Dr Raffaele Conte, PhD, FIScT, RSci is the director and responsible chemist of the toxicology laboratory of AMES polydiagnostic centre in Casalnuovo, Italy and a research associate of the Research Institute on Terrestrial Ecosystems of the Italian National Research Council (CNR) in Naples, Italy. Apart from his working activity on the detection of drugs and metabolites in biological matrices, his research interests are focused on the design of novel delivery platforms able to minimize degradation, prevent undesirable side effects, and increase bioavailability of drugs or genetic materials. He also follows different projects as an independent researcher and has authored several research papers. In 2018 he obtained a PhD in clinical and experimental medicine at the University of Naples “Federico II” with a project on the synthesis of polymeric fillers with anti-biofilm activity for dental composites. His email address is raffaele.conte86@tiscali.it
References
-
Zhu N, Zhang D, Wang W, et al. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine 382: 727-733.
-
Chan JF-W, Yuan S, Kok K-H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet 395: 514-523.
-
Wu JT, Leung K, Bushman M, et al. (2020) Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nature Medicine 26: 506-510.
-
Charlton CL, Babady E (2019) Practical Guidance for Clinical Microbiology Laboratories: Viruses Causing Acute Respiratory Tract Infections. 32.
-
Falsey AR, Formica MA, Walsh EE (2012) Simple method for combining sputum and nasal samples for virus detection by reverse transcriptase PCR. Journal of clinical microbiology 50: 2835-2835.
-
Wang W, Xu Y, Gao R, et al. (2020) Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA.
-
Cheng PK, Wong DA, Tong LK, et al. (2004) Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet 363: 1699-1700.
-
Briese T, Mishra N, Jain K, et al. (2014) Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio 5: e01146-01114.
-
Chen L, Liu W, Zhang Q (2020) RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. 9: 313-319.
-
Mahony JB, Richardson S (2005) Molecular diagnosis of severe acute respiratory syndrome: the state of the art. The Journal of molecular diagnostics : JMD 7: 551-559.
-
Zlateva KT, Coenjaerts FEJ, Crusio KM, et al. (2013) No novel coronaviruses identified in a large collection of human nasopharyngeal specimens using family-wide CODEHOP-based primers. Archives of virology 158: 251-255.
-
Woo PC, Lau SK, Chu CM, et al. (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79: 884-895.
-
Kuypers J, Martin ET, Heugel J, et al. (2007) Clinical Disease in Children Associated With Newly Described Coronavirus Subtypes. Pediatrics 119: e70-e76.
-
Babady NE, England MR, Jurcic Smith KL, et al. (2018) Multicenter Evaluation of the ePlex Respiratory Pathogen Panel for the Detection of Viral and Bacterial Respiratory Tract Pathogens in Nasopharyngeal Swabs. 56.
-
Babady NE, Mead P, Stiles J, et al. (2012) Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol 50: 2282-2288.
-
Holshue ML, DeBolt C, Lindquist S, et al. (2020) First Case of 2019 Novel Coronavirus in the United States. New England Journal of Medicine 382: 929-936.
